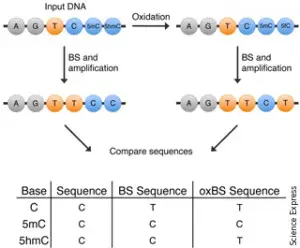
Cytosine methylation can significantly modify temporal and spatial gene expression and chromatin remodeling. Bisulfite Sequencing, BS, leverages the power of massively parallel sequencing and genome-wide analysis to provide a comprehensive view of methylation patterns at single-base resolution across the genome. Treatment of DNA with bisulfite converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. Thus, bisulphite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues.
Advantages of Bisulfite Sequencing
- Discover methylation patterns of all CpG, CHH, and CHG regions across the entire genome
- Capture full sample diversity with small amounts of DNA
- View methylation at practically every cytosine in the genome across most species
Bisulfite Sequencing Workflow
First, we perform a quality control step in which we measure the quantity and quality of the DNA provided to us by our customers. Only when the provided genomic DNA is of sufficient quality and quantity will we proceed with the next steps. In case the DNA provided to us is of insufficient quality and/or quantity, we will contact the customer and discuss how to proceed.
In the next steps the DNA samples are bisulfite treated before library preparation. Sequencing libraries are again quality checked. The library preparation and sequencing are then performed using one of our Illumina sequencers. The technical quality of the sequencing run is monitored in real time.
Sequencing data can be transferred to you via the Illumina BaseSpace platform or our own server (FTP download).
For more information, please feel free to contact us.