By combining chromatin immunoprecipitation (ChIP) assays with sequencing, ChIP sequencing (ChIP-Seq) is a powerful method for identifying genome-wide DNA binding sites for transcription factors and other proteins. The application of next-generation sequencing (NGS) to ChIP has revealed insights into gene regulation events that play a role in various diseases and biological pathways, such as development and cancer progression. ChIP-Seq enables thorough examination of the interactions between proteins and nucleic acids on a genome-wide scale.
Common targets for ChIP-seq
- Transcription factors
- Histone methylation
- Histone acetylation
- Histone ubiquitination
- …
Advantages of ChIP-Seq
- Captures DNA targets for transcription factors or histone modifications across the entire genome of any organism
- Defines transcription factor binding sites
- Reveals gene regulatory networks in combination with RNA sequencing and methylation analysis
- Offers compatibility with various input DNA samples
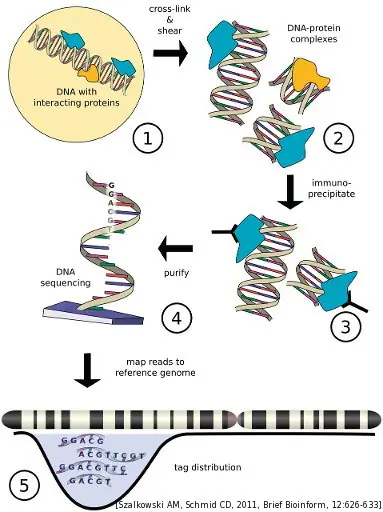
ChIP-seq workflow
Following ChIP protocols, DNA-bound protein, such as histones, is immunoprecipitated using a specific antibody. The bound DNA is then coprecipitated, purified, and sequenced on of our Illumina sequencers. The resulting sequencing reads are mapped back to the reference genome. This will effectively give you an idea of where your target protein-DNA interactions occur throughout the entire genome. The technical quality of the sequencing run is monitored in real time.
Sequencing data can be transferred to you via the Illumina BaseSpace platform or our own server (FTP download).
For more information, please contact us.