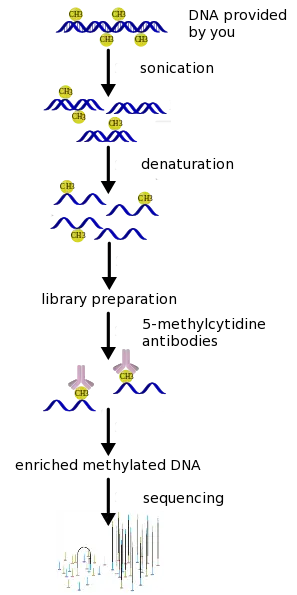
(h)MeDIP-seq combines methylated DNA immunoprecipitation (MeDIP) with massively parallel DNA sequencing. It involves antibodies directed against mC/mCG to precipitate methylated DNA fragments. (h)MeDIP is able to detect methylated cytosines in both mC and mCG contexts. Combining MeDIP with next generation sequencing provides high-quality methylomes at typically 100-300bp resolution at costs comparable to other capture-based techniques.
Advantages of (h)MeDIP sequencing
- True genome-wide
- Focus on less-densily methylated regions
- Approximate basepair resolution
- Applicable to all mammalian samples and tissue types
(h)MeDIP Sequencing Workflow
First, we perform a quality control step in which we measure the quantity and quality of the DNA provided to us by our customers. Only when the provided genomic DNA is of sufficient quality and quantity will we proceed with the next steps. In case the DNA provided to us is of insufficient quality and/or quantity, we will contact the customer and discuss how to proceed.
In a second step we perform a random fragmentation of the genomic DNA samples followed by size selection of the resulting fragments. This is followed by a second quality control step in which the resulting DNA concentration is determined and the DNA is checked for degradation.
On the DNA fragments a library will be prepped by ligating adaptors. The resulting DNA fragments will then be separated by performing MeDIP-capture during which all fragments containing methylation are withheld, while fragments that do not contain methylation are discarded. Once again the DNA concentration will be determined during as a quality control step.
The library preparation and sequencing are then performed using one of our Illumina sequencers. The technical quality of the sequencing run is monitored in real time.
Sequencing data can be transferred to you via the Illumina BaseSpace platform or our own server (FTP download).
For more information, please feel free to contact us.